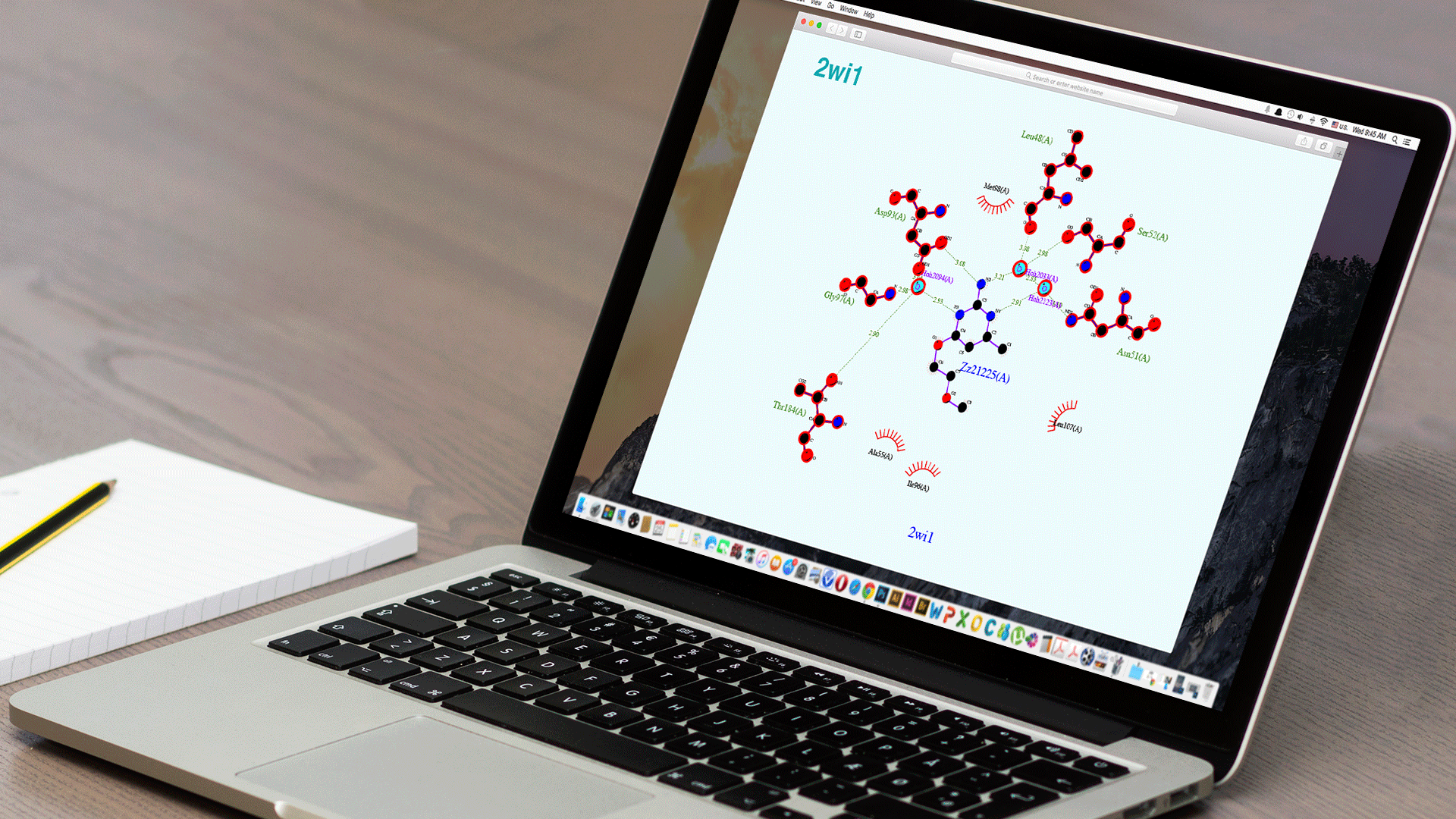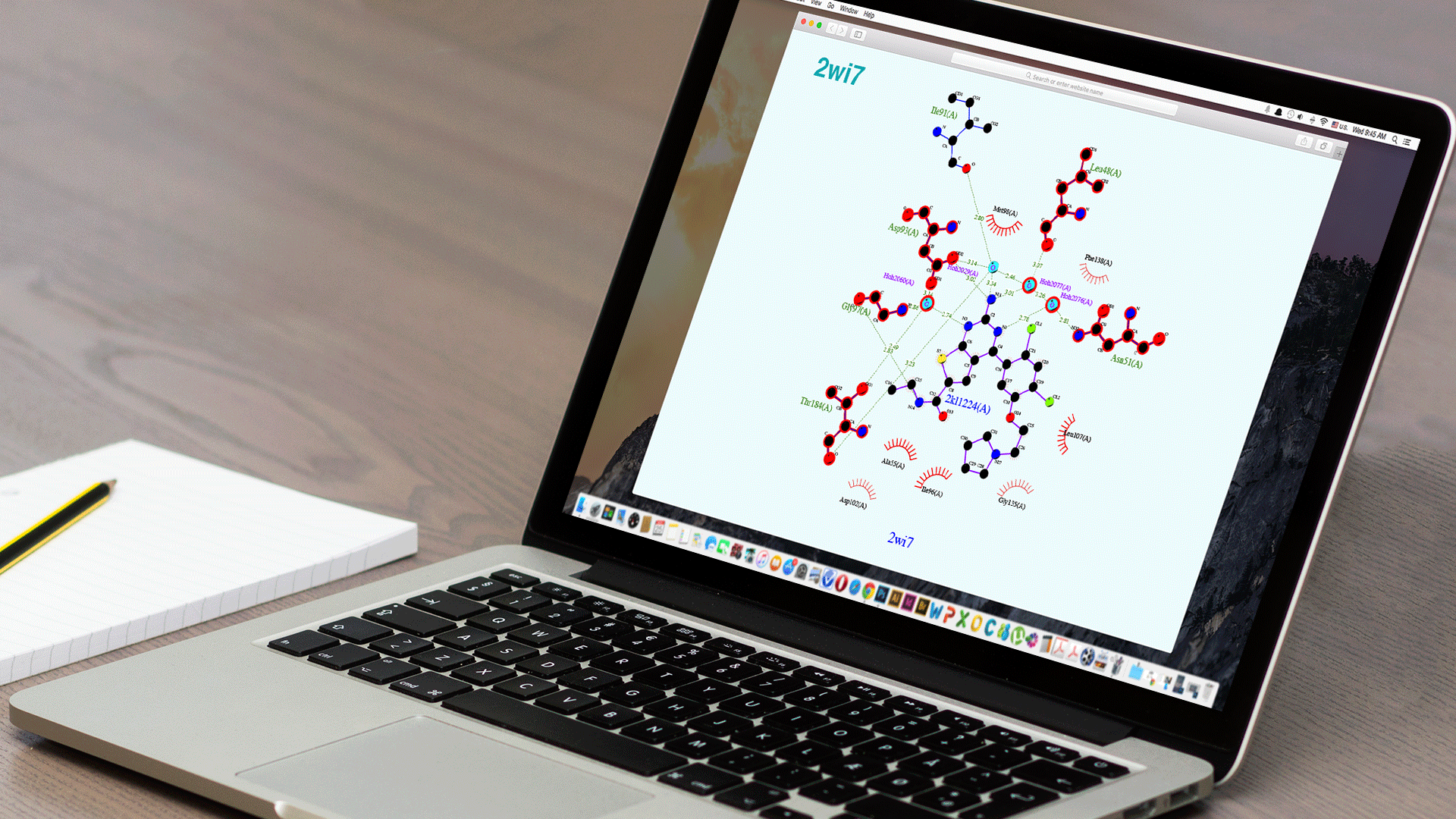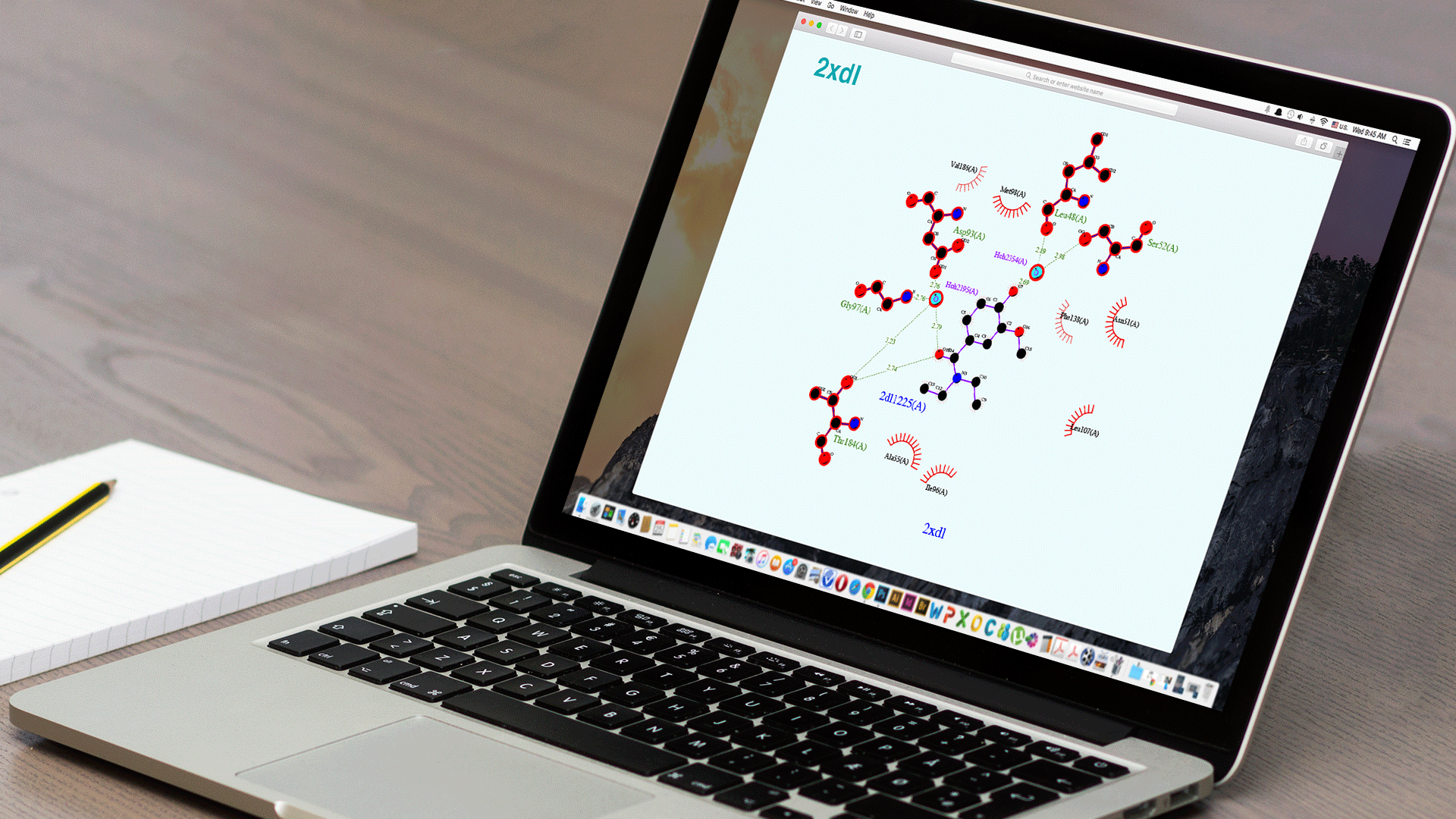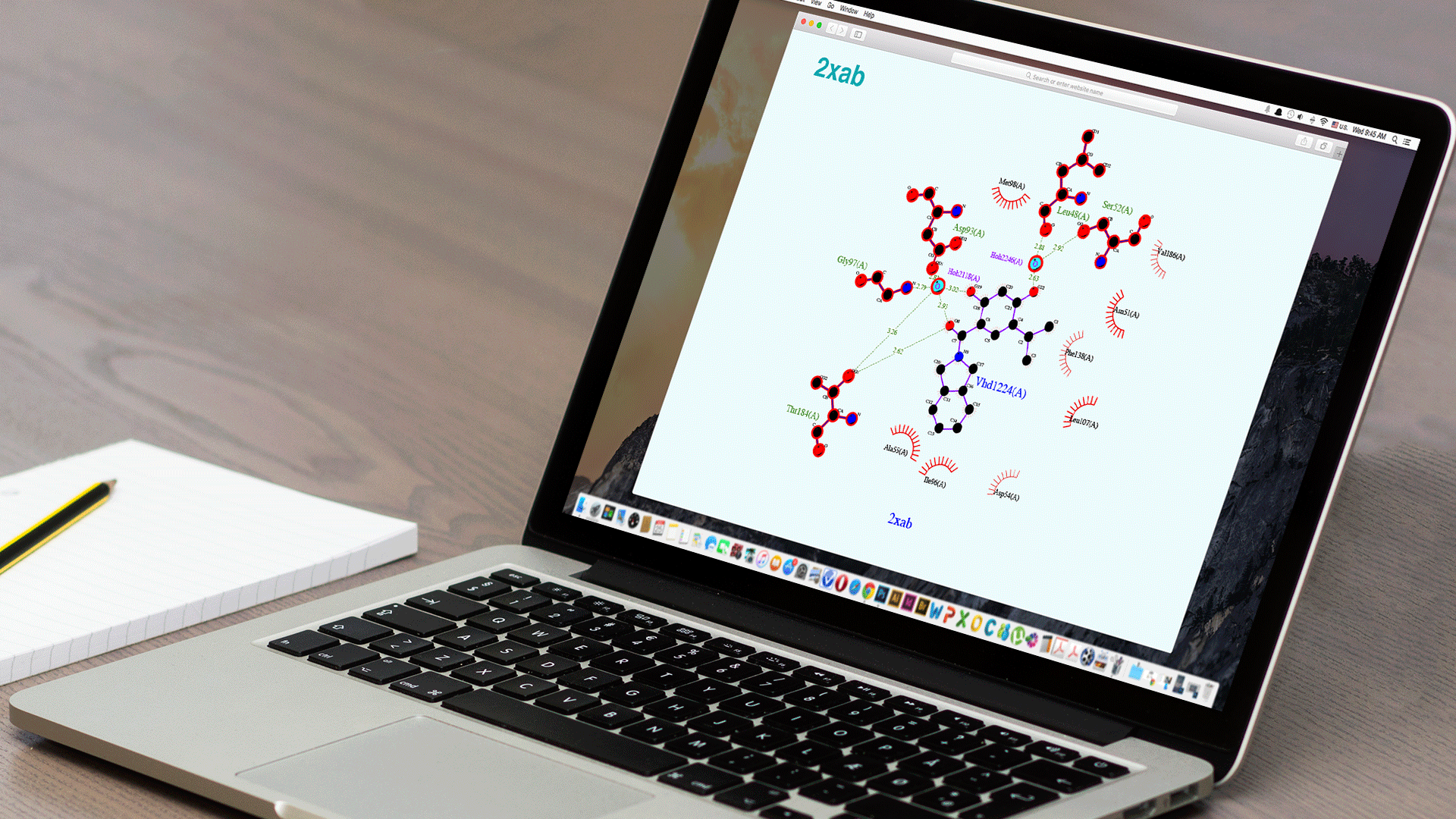
Ligplot+
The original software for 2D representations of Ligand-Binding site interactions.
Ligplot+ is the original software for 2D representations of Ligand-Binding site interactions.
With the latest version you can also flexibly compare:
Same protein with different ligands (specificity)
Homologous proteins with the same ligand (selectivity)
Homologous proteins with different ligands
Be guided in the analysis of selectivity and specificity through with detailed automated comparisons of homologous binding site interactions. Even analyse distant homologues with comparable binding sites by uploading a structure based sequence alignment.
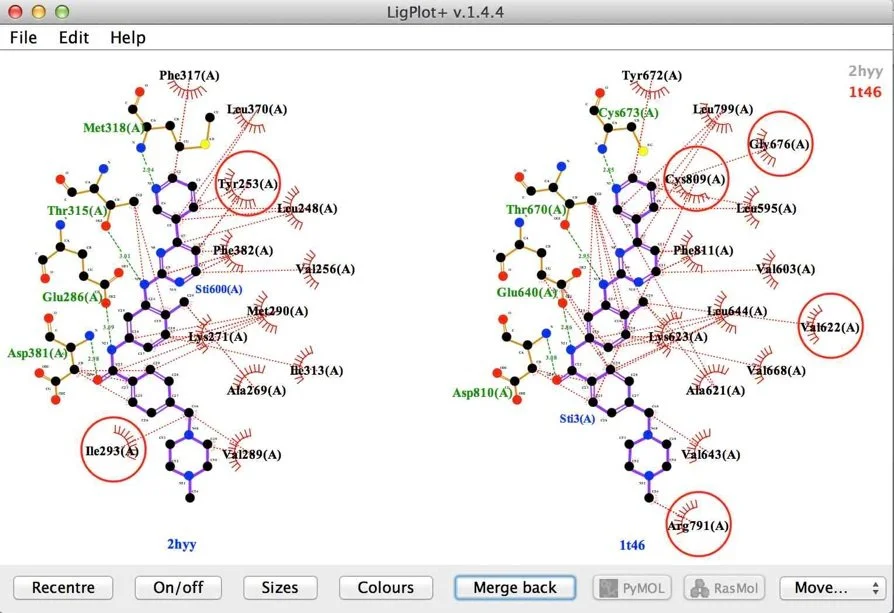
Homologous Proteins, Same Ligand
Analysis of Imatinib with homologues c-abl (2hyy) and c-kit (1t46).
The two sequences have 36% sequence identity. Conserved interactions predominate so unconserved positions have been highlighted
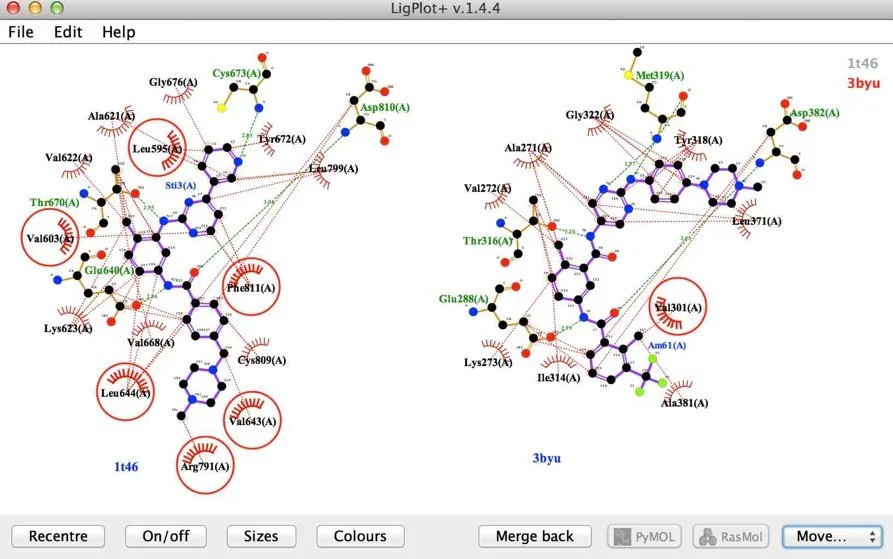
Homologous Proteins, Different Ligands
Imatinib complex with homologue c-kit (2hyy) and amino pyrimidine reverse amide with lck (1t46).
37% sequence identity and 0.85 Tanimoto coefficient. The large difference in binding observed in 3D is reflected by Ligplot+. Again Ligplot+ highlights dis-similar interactions in this figure.